Sugar signalling
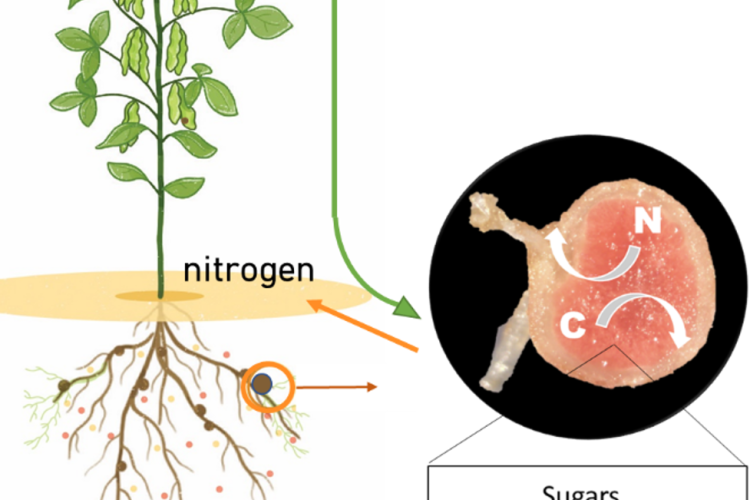
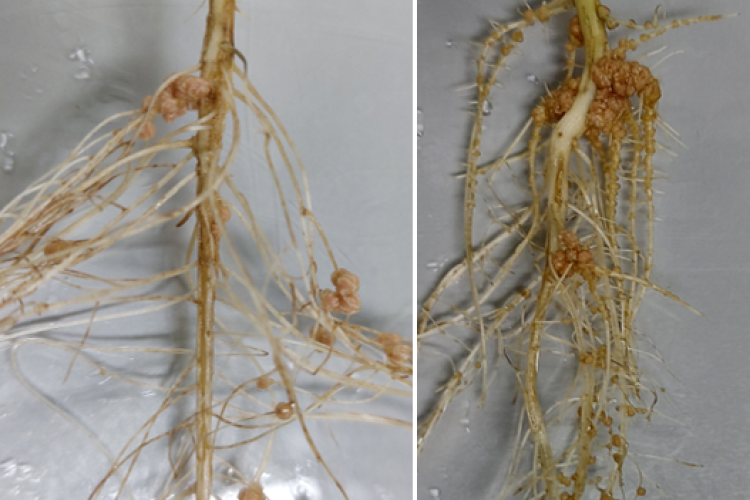
Because of the ever increasing food and feed demand and the need for reducing global greenhouse gas emissions, the world is in desperate need for sustainable solutions in agriculture. Legume plants are an excellent solution as they can be used in intercropping systems, as green manure, and as alternative protein sources. Indeed, they contain high protein levels due to their engagement with endosymbiotic diazotrophic bacteria. These so-called rhizobia biologically fix nitrogen (N) into plant-accessible forms in newly formed root structures called nodules. However, to fix N, these rhizobia need carbon (C), delivered by the plant in the form of sugars. Sugars are generated during photosynthesis in leaves and transported to non-photosynthetic sink tissues such as flowers, roots, and nodules. Surprisingly, even though the important role of sugar metabolism and signalling in plant growth is well-established and successful nodulation strongly depends on a correct exchange of C/N between both symbiotic partners, today, we still do not exactly know how sugars are metabolized, sensed, and regulated locally inside legume nodules at the cellular level.
Our research tries to answer the following topics:
1. Sugar Sensing
We aim to resolve which, where, how and when the three major conserved regulators TARGET OF RAPAMYCIN (TOR), SNF1-RELATED PROTEIN KINASE1 (SnRK1), and HEXOKINASE1 (HXK1) regulate soybean nodulation, using the cutting-edge single-cell, spatial, and multi-omics (interactomics, transcriptomics and metabolomics) approaches, allowing the integration of the cellular complexity of a nodule.
2. Sugar metabolism and transport
We study how sugar metabolism and transport is modulated inside soybean nodules by combining single nuclei transcriptomics, spatial transcriptomics and spatial metabolomics on young immature, mature and senescent nodules.
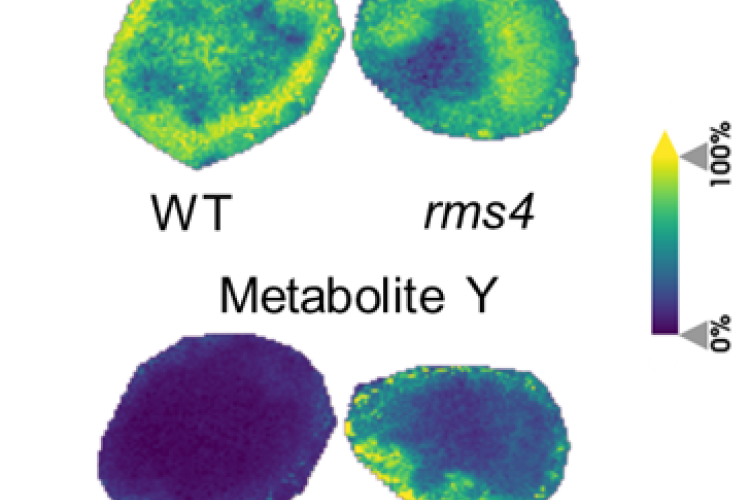
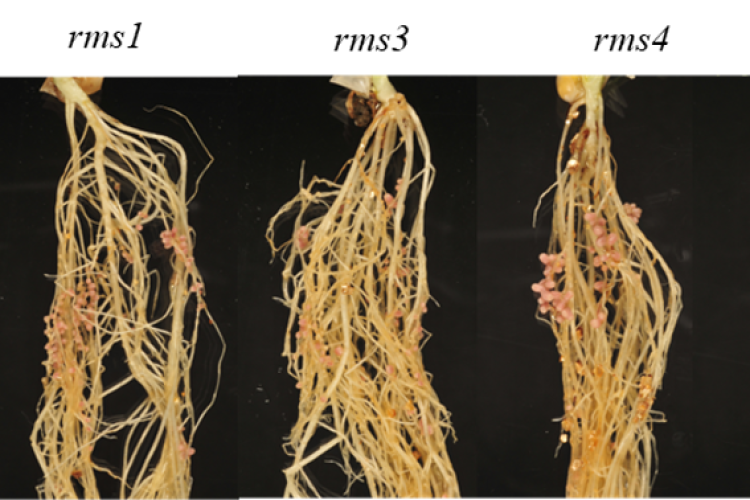
3. Strigolactones in pea nodule development
By means of phenotypical, molecular, and (spatial) metabolomics analyses of nodules, we investigate the involvement of the plant hormones strigolactones in pea nodule development and how this process is connected to the energy (sugar) availability in the plant.
Autoregulation of nodulation
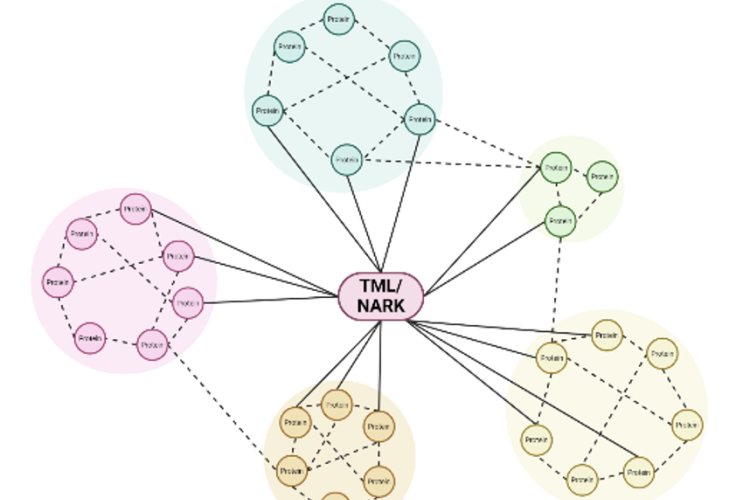
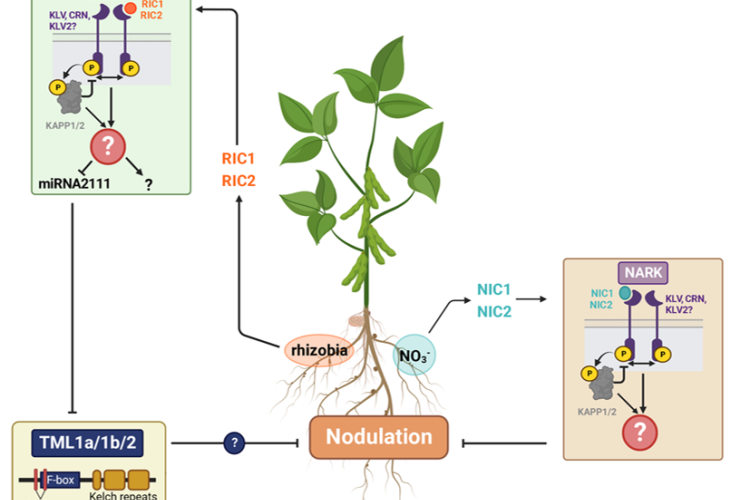
Legume plants have evolved regulatory pathways to control nodule numbers and obtain an optimal balance between the benefits (N) and costs (C) of this interaction. A negative feedback mechanism that controls nodule numbers systemically is activated upon interaction with rhizobia, this is called autoregulation of nodulation (AON). AON involves the transport of CLE peptides from root to shoot, where they are perceived by a leucine-rich repeat receptor like kinase (LRR-RLK), called NODULE AUTOREGULATION RECEPTOR KINASE (NARK) in soybean and SUPER NUMERARY NODULES (SUNN) in Medicago truncatula. Downstream signalling results in reduced shoot-to-root transport of miR2111, which allows accumulation of Too Much Love (TML) in roots. TML is a nodulation-inhibiting F-box protein, which is expected to ubiquitinate target proteins. In addition to AON, nitrate availability also inhibits nodulation via CLE peptide signalling. Although NARK and TML are key players in the AON pathway, their downstream signalling and targets remain unresolved. That is why our research focuses on the following topics:
1. AON in soybean
We aim to elucidate the regulation of nodulation pathway in soybean plants using a proteomics approach. Time course experiments will allow to characterize dynamic proteome changes in soybean roots and leaves in response to rhizobia. In particular, phospho- and ubiproteomics are applied to obtain more insights in downstream NARK and TML signalling respectively. Furthermore, this will be complemented by TurboID, allowing to identify putative interactors of known players in the AON pathway.
2. AON in Medicago
By performing proteomics in CLE13-overexpressing mutants of Medicago truncatula, we are trying to elucidate which proteins are involved in the AON pathway. These results will be further analysed using transcriptional approaches, GUS staining and phenotypical analyses in Medicago truncatula (hairy root) mutants.